Developing novel cancer treatment methods

DNA methylation is one of the epigenetic modifications that allow cancer cells to resist any form of treatment. Epigenetic changes control the phenotypic plasticity of cancer cells, allowing them to evade practically every therapy, such as chemotherapy, radiation, surgery, etc. In the article “SERS-Based Evaluation of the DNA Methylation Pattern Associated With Progression in Clonal Leukemogenesis of Down Syndrome”, published in Frontiers in Bioengineering and Biotechnology, scientists try to understand the epigenetic mechanisms underlying cancer genesis and progression for developing novel cancer treatment methods.
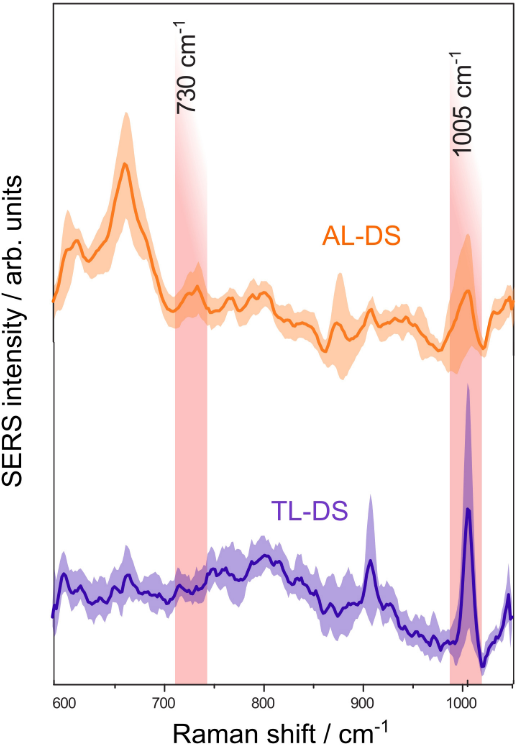
SERS is an ideal tool for studying acute leukemia associated with Down syndrome (AL-DS) pathology because the DNA of patients with AL-DS has a distinctive global hypomethylation. Researchers carried out measurements in a liquid drop of silver colloidal solution. To determine the difference between the two types of genomic DNA (transient leukemia associated with Down syndrome (TL-DS) versus AL-DS), the authors compared the area under the 1005 cm−1 band (see Fig.1). The average area was lower for AL-DS patients than for TL-DS. Therefore, compared to DNA from TL-DS patients, AL-DS patients' DNA is relatively hypomethylated. These findings suggest that DNA methylation decreases as cells develop from normal to pre-leukemia to full-blown acute leukemia. Thereby, SERS is a potential technique for analyzing DNA.
Figure 1. Average SERS spectra of DNA from patients with acute leukemia associated with Down syndrome (AL-DS) and transient leukemia associated with Down syndrome (TL-DS). Adopted from here.